John Parsch - Evolutionary and Functional Genomics
In general, we are interested in understanding the molecular basis of adaptation. We study the evolution of genes and gene expression using the fruit fly Drosophila melanogaster as a model system. Current projects focus on variation in gene expression between populations and sexes, as well as the population genetic and functional analysis of gene regulatory elements. Our research can be divided into two major areas:
Evolutionary and Population Genetics of Gene Expression
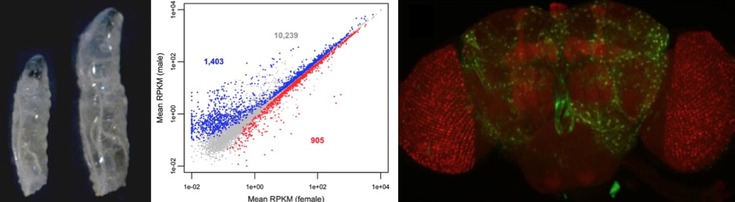
Differences in gene expression are thought to underlie many of the phenotypic differences between species and populations. Transcriptomic technologies, such as high-throughput RNA sequencing (RNA-seq), have made it possible to identify the genes that differ in expression between species or vary in expression among individuals of the same species. Such studies have revealed that there is considerable expression divergence between closely related species, as well as abundant expression variation within species. Furthermore, there are extensive differences in gene expression between males and females of the same species. A current challenge in evolutionary genetics is to identify the specific genetic changes responsible for differences in gene expression and to determine how these changes impact an organism's fitness.
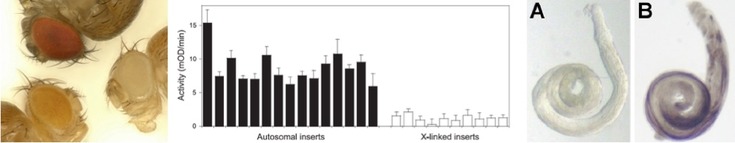
We are studying gene expression variation within and between populations of the fruit fly Drosophila melanogaster, a species that has its origin in sub-Saharan Africa and only relatively recently has become a successful colonizer of other word-wide habitats. In order to better understand the role of gene regulatory changes in adaptation, we conduct both genome-wide and candidate-gene studies using flies from ancestral and derived populations. The long-term goal is to identify specific genetic polymorphisms that underlie gene expression divergence and to determine the population genetic mechanisms that are responsible for maintaining them in natural populations. A further goal is to determine the effect that variation in gene expression has on an organismal phenotype that may be subject to natural selection.
Evolution and Expression of Sex Chromosomes
Similar to humans, Drosophila have chromosomal sex determination, with females having two X chromosomes and males having one X and one Y chromosome. The male-specific Y chromosome is highly degenerated and contains very few genes. The X chromosome, in contrast, contains many genes (about 17% of the genes in the Drosophila genome) that are expressed in both sexes. The difference in copy number between males and females makes the X chromosome subject to unique evolutionary forces and gene regulatory mechanisms. For example, in males, the expression of genes on the X chromosome is increased in order to compensate for it being present in only one copy. This process is known as dosage compensation. Furthermore, it has been found that expression of the X chromosome is suppressed in the male germline.
We are currently investigating the extent of X-chromosome suppression (what type of genes and which tissues are affected?), as well as the mechanisms responsible for it. We are also studying variation in dosage compensation among tissues and how the proximity of a gene to a binding site of the dosage compensation complex (DCC) influences its expression level in males.
Representative publications:
Ramnarine TJS, Grath S, Parsch J. 2022. Natural variation in the transcriptional response of Drosophila melanogaster to oxidative stress. G3 (Bethesda) 12: jkab366.
Full online text
Glaser-Schmitt A, Wittmann MJ, Ramnarine TJS, Parsch J. 2021. Sexual antagonism, temporally fluctuating selection, and variable dominance affect a regulatory polymorphism in Drosophila melanogaster. Molecular Biology and Evolution 38: 4891-4907.
Full online text
Glaser-Schmitt A., and J. Parsch (2018) Functional characterization of adaptive variation within a cis-regulatory element influencing Drosophila melanogaster growth. PLoS Biol 16: e2004538.
Full online text
Catalán A., A. Glaser-Schmitt, E. Argyridou, P. Duchen, and J. Parsch (2016) An indel polymorphism in the MtnA 3' untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 12(4):e1005987.
Full online text
Huylmans, A. K., and J. Parsch (2015) Variation in the X:autosome distribution of male-biased genes among Drosophila melanogaster tissues and its relationship with dosage compensation. Genome Biol. Evol. 7: 1960-1971.
Full online text
Kemkemer, C., A. Catalán, and J. Parsch (2014) Escaping the X chromosome leads to increased gene expression in the male germline of Drosophila melanogaster. Heredity 112: 149-155.
Full online text

