Go to the "Download and Documentation" section to download the R-Files for the processing of the TreeTime output.
Download and unzip the *.r files in the same directory as the output files.
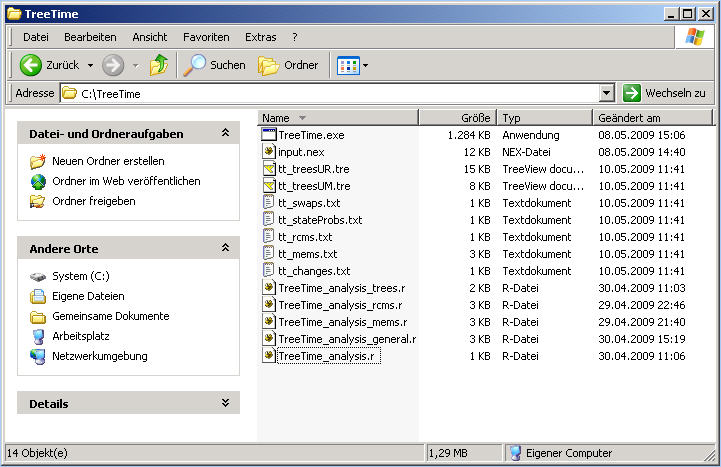
Now your folder should look similar to this:

Navigate to the output directory and execute R.
Make sure that the R-command getwd() returns the output directory containing the TreeTime output and the unzipped R-files.
Otherwise use the R-command setwd("path") to navigate to the correct directory.
Type the R-command source("TreeTime_analysis.r").
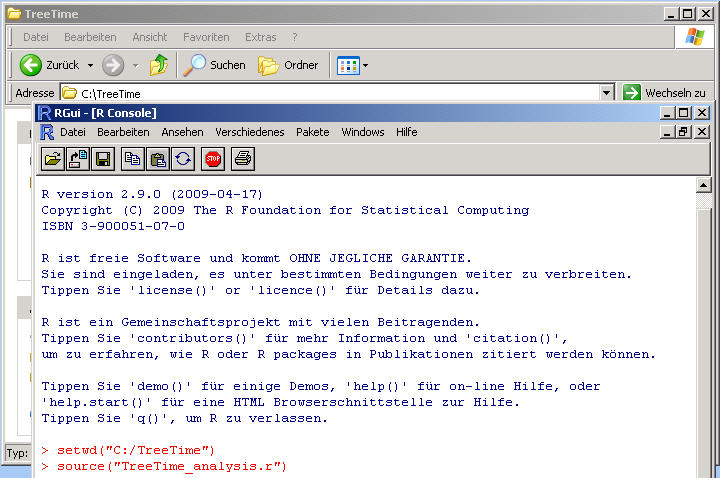
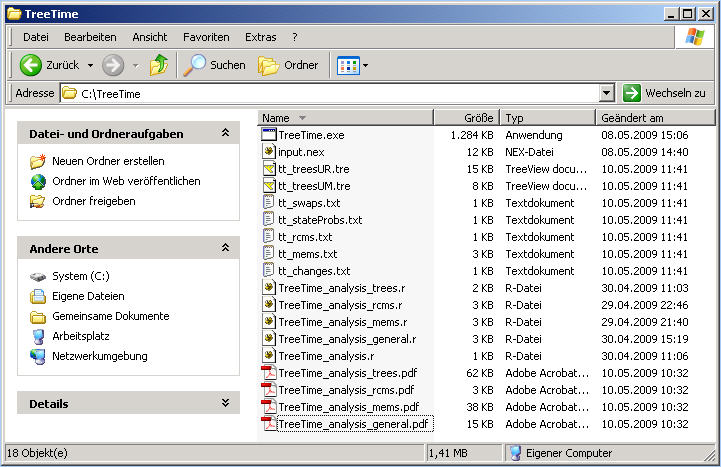
After the R-command source("TreeTime_analysis.r") has finished,
you will find 4 *.pdf files in the directory that contain the summary of the the analysis, prints different consensus trees,
shows the model parameters to monitor convergence, etc..
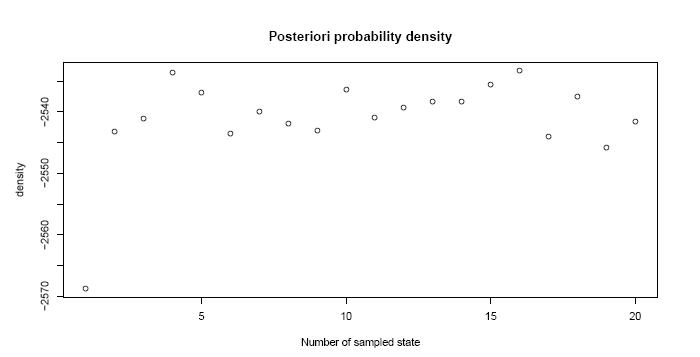
The file TreeTime_analysis_general.pdf contains data to the mcmc-method.
For example the posterior density of the sampled states...
the Likelihood of the sampled states...
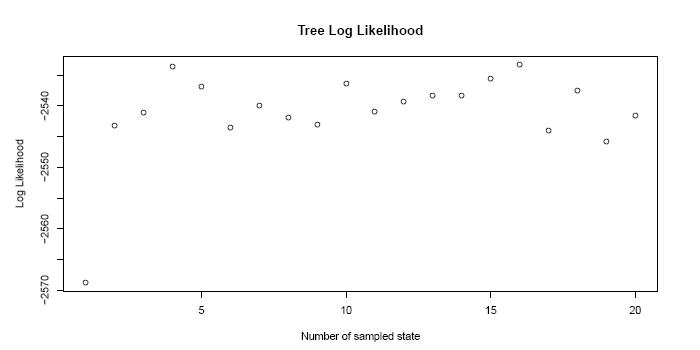
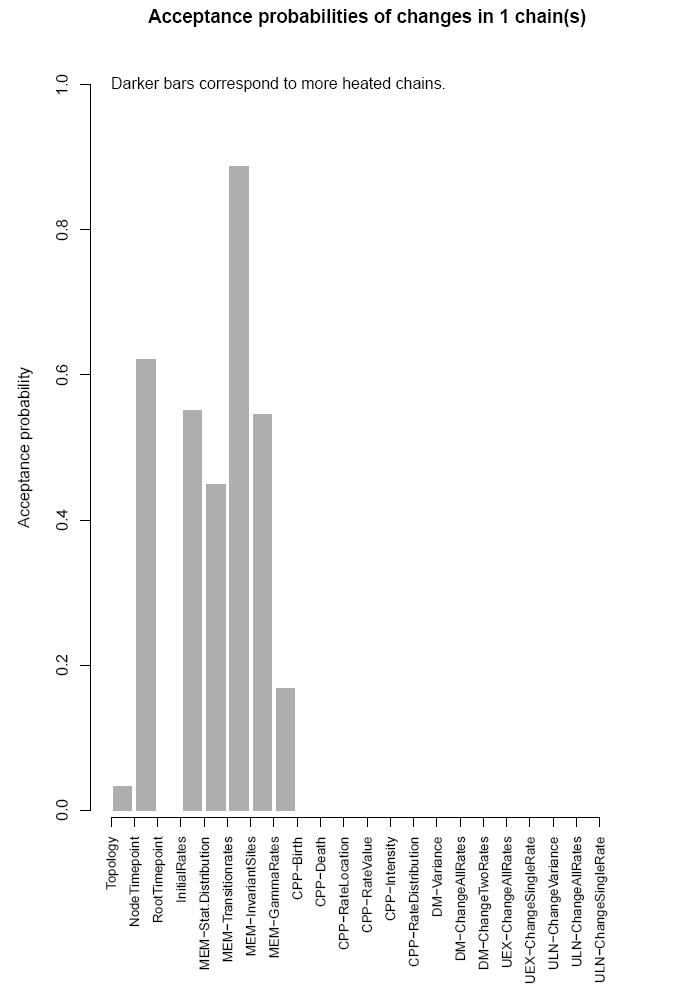
and the acceptance probabilities of the proposed moves in the mcmc-algorithm.
If you run several chains, the brightness of the bars in the plot will encode the different chains.
The file TreeTime_analysis_mems.pdf contains data to the molecular evolution models.
For example the proportion of invariant sites in the sampled states...
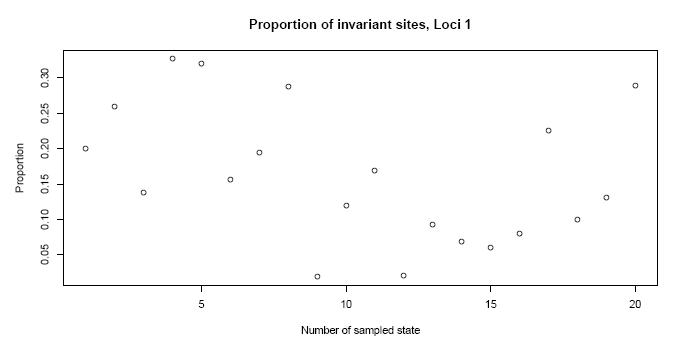
the stationary distribution of the nucleotides...
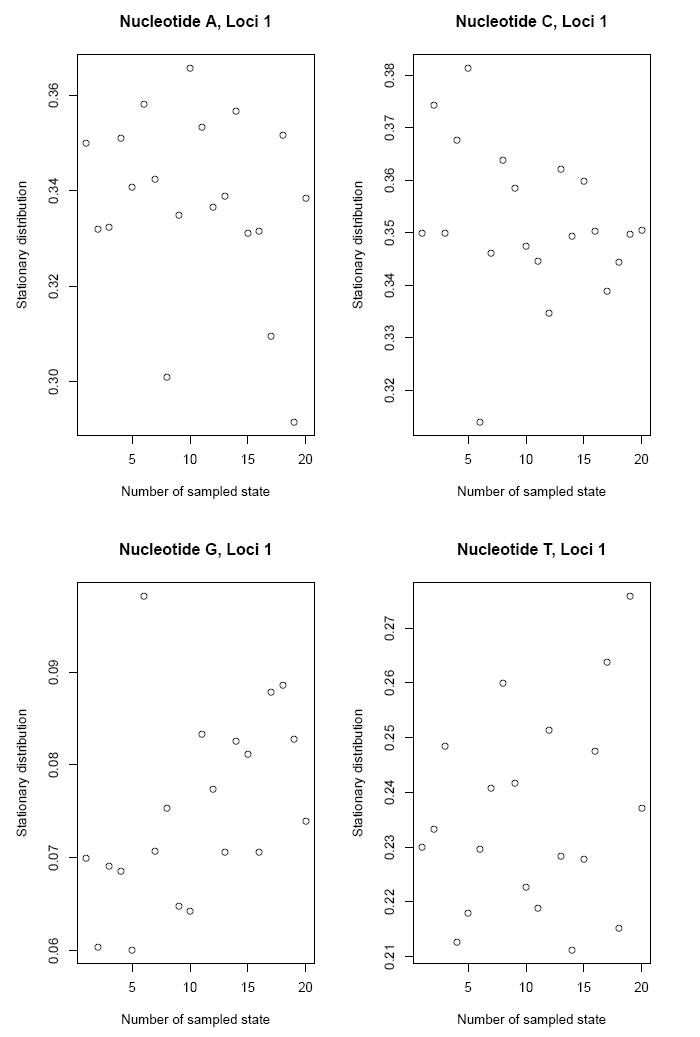
and the mutation rates of the nucleotides.
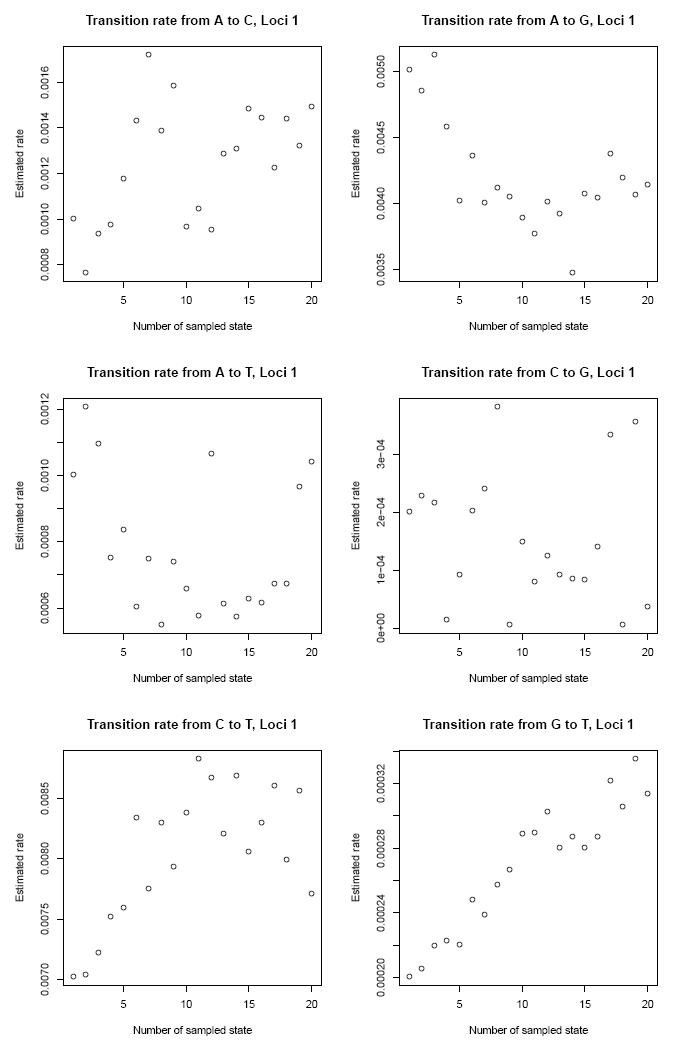
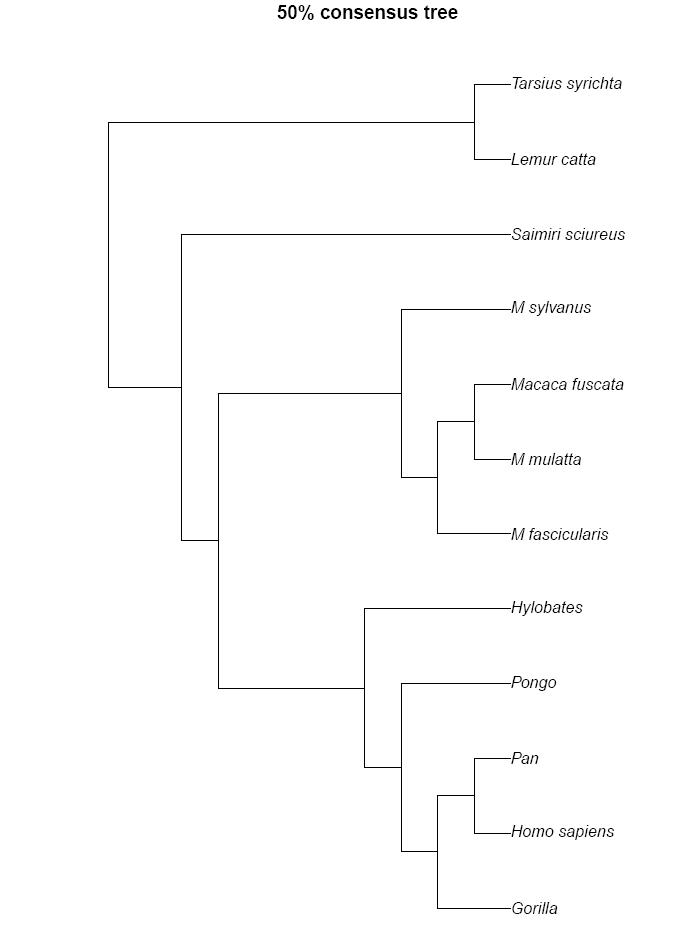
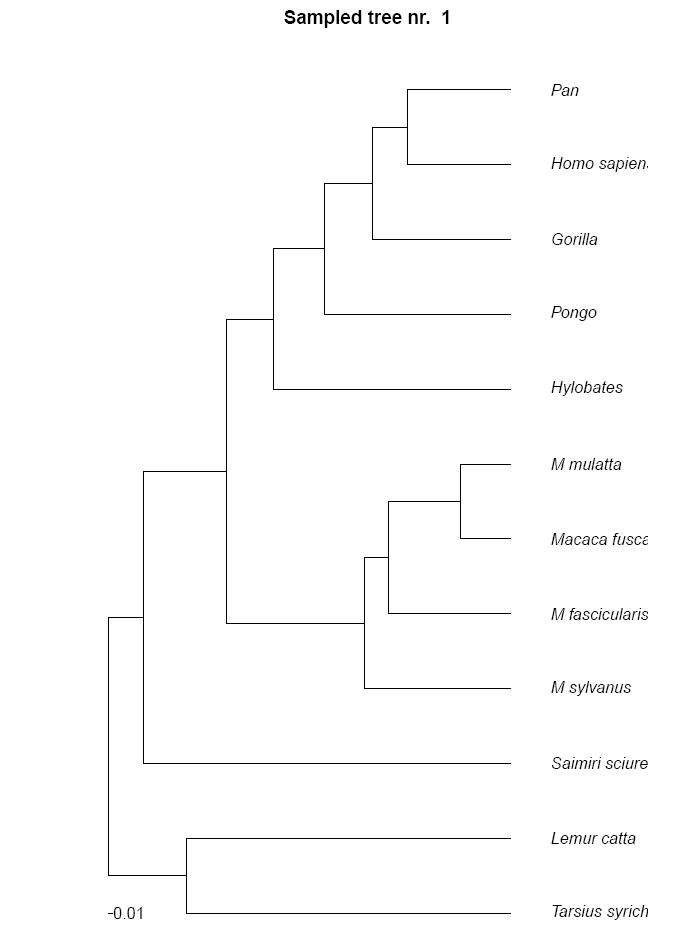
The file TreeTime_analysis_trees.pdf contains for example
the 50% consensus tree...
and the trees of the sampled states.
If you run the analysis with a rate change model, the file TreeTime_analysis_rcms.pdf contains the parameters of the used model.
IMPRESSUM, 10 May 2009, Lin Himmelmann